Microchem’s microbial genetic sequencing accurately identifies an organism with the help of phylogenetics.
The Science Behind Phylogeny:
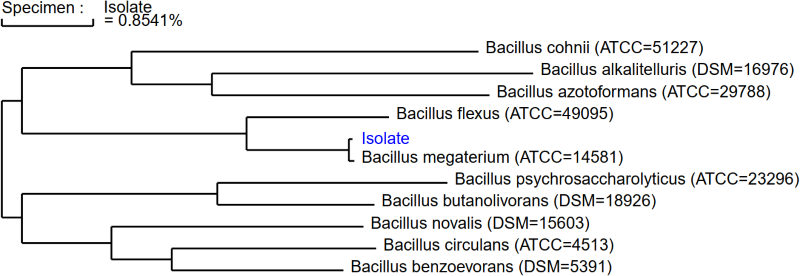
Phylogeny is the method of understanding evolutionary relationships by comparing genetic information of organisms. Often, a phylogenetic tree is used to help pinpoint species of similar descent, where each branch resembles a change in physical or genetic variance. By comparing a multitude of known genetic species, our lab identifies organisms with MicroSEQ ID: a database that utilizes a phylogenetic tree to display isolate reports.
The common genetic marker used for bacterial identification is the 16S rRNA gene sequence. When molecularly isolating bacteria, this gene is used for a multitude of reasons. One of those reasons being the gene’s presence in almost all bacterium and still maintaining the same function. Because of this, scientists view this gene as a sort of timestamp. When there are variants within the gene sequence, they resemble different bacterial lineages. Although the function of the gene has had no significant evolutionary change over time, the 16S rRNA gene sequence still has great variability and uniqueness among different organisms, making a microbe’s identity easily known when comparing the isolate’s DNA to a large database. The 16S rRNA gene is also highly conserved, allowing universal primers to pair and sequence the gene both forward and backward. A highly conserved gene maximizes the number of nucleotides read during sequencing. The unique molecular structure the 16s RNA gene encompasses makes for the best DNA analysis when conducting a phylogenetic tree for bacteria. Having similar attributes, the D2 large-subunit rDNA region is used for identifying fungal contaminants.
Phylogeny and MicroSEQ ID:
Often referred to as the gold standard of microbial identification, Microchem takes a molecular approach to identify contamination points. Our lab locates and amplifies the 16S rRNA gene for bacterial samples and the LSU D2 region for fungi, regions often used to compare phylogenetic relationships among organisms. As the final step in our microbial identification process, MicroSEQ ID comparatively analyzes the DNA sequence results to a database of over 20,000 validated species. Having a large collection of genetically known taxa makes finding the microbial’s identity seamless. The genetic sequence is ranked in similarity to the database’s organisms and displays the results on a phylogenetic tree. The phylogenetic tree allows our lab to report, with confidence, the correct identity of your isolate.