Microchem Laboratory uses a state-of-the-art genetic sequencer to identify unknown bacteria and fungi. This article explains the science behind the service.
First Step: PCR Amplification
Starting with DNA extracted from the environmental isolate, Polymerase Chain Reaction (PCR) is performed to amplify just a small region of the DNA used to identify the isolate. For bacteria, the 16S ribosomal RNA (rRNA) gene sequence is used. The 16S rRNA sequence is common across all bacterial species, and can be used to study the phylogenetic relationship among bacteria. Typically for MicroSEQ ID workloads, the first 500 nucleotide bases (the A, C, G, and T of DNA) of the sequence are sufficient to identify the majority of species. For fungi, the D2 region of the larger rRNA molecule as a part of the ribosome is most often used for identification.
Within these amplified regions are sections that are conserved – the same for all – and some that are hypervariable, or widely different depending on the species. The conserved region acts as a framework that can be used to amplify the DNA, and the hypervariable regions allow for fingerprinting of each species. You may notice that both bacteria and fungi are identified using sequences from the ribosome – these universal biomarkers are common to all life, yet have minute differences allowing reliable identification of the species.
Next: Cycle Sequencing
A second round of PCR is performed to add individual fluorescent nucleotides to the amplified DNA, with each type of nucleotide having a unique color. The colored nucleotides are only added to the end of each new DNA segment, and the amplification ends at a variety of lengths of DNA to provide a spectrum of pieces. As an example, a starting DNA sequence of “AATCGA” would yield all of the following after cycle sequencing: “AATCGA”, “AATCG”, “AATC”, “AAT”, “AA”, “A” – with each piece labeled at the end with a colored base. This is a form of Sanger sequencing, and the different sized pieces will be used to read the final sequence.
Next: Capillary Electrophoresis
The samples are injected into thin, long capillaries containing a gel that slows the pieces of the amplified DNA. A strong electric field is run across the capillary, bringing the negatively charged DNA to the other end. Larger pieces of DNA run through the capillary slower, and small pieces faster. This is known as “electrophoresis”, and is the key method by which the DNA sequence is identified. At the end of the capillary is a camera detector which can identify each of the 4 colored nucleotides. Using the “AATCGA” sequence example from before, first the initial “A” would reach the detector, followed by “AA”, then “AAT”. The detector only sees the color from the last nucleotide of each segment, so it would read first “A”, then another “A, then a “T”, and so on. By this method, the genetic code is slowly built up until the final sequence is put together.
Finally: Library Search and Phylogenetic Analysis
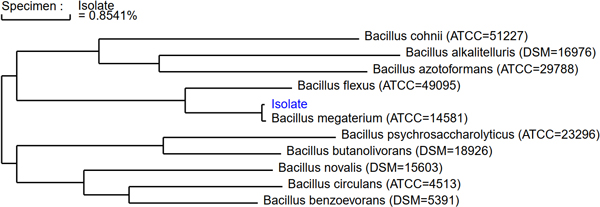
The detected DNA sequence is finally compared to a validated library of other the rRNA sequences from other bacteria or fungi. Microchem uses MicroSEQ ID validated libraries, containing over 20,000 species. The sequence is compared to all other species in the library, building up a phylogenetic tree – a graph showing the closest relations of the starting isolate to other known species. A percentage similarity of the new sequence to these closest related species is given, and a 99% or greater sequence similarity is used to classify the isolate as a specific species.